-Search query
-Search result
Showing all 35 items for (author: zivanov & j)

EMDB-16183: 
In situ structure of the Caulobacter crescentus S-layer
Method: subtomogram averaging / : von Kuegelgen A, Bharat T

EMDB-16207: 
HIV-1 CA-SP1 subtomogram average with Relion4 from EMPIAR-10164 dataset, 3.2 A from 5 tomograms
Method: subtomogram averaging / : Ke Z, Briggs JAG, Scheres SHW

EMDB-16209: 
HIV-1 CA-SP1 subtomogram average with Relion4 from EMPIAR-10164 dataset, 3.0 A from the full dataset
Method: subtomogram averaging / : Ke Z, Briggs JAG, Scheres SHW

PDB-8bqe: 
In situ structure of the Caulobacter crescentus S-layer
Method: subtomogram averaging / : von Kuegelgen A, Bharat T

EMDB-15949: 
COPII inner coat reprocessed with relion4.0
Method: subtomogram averaging / : Zanetti G, Pyle E

EMDB-11493: 
Subtomogram averaging of SARS-CoV-2 Spike Protein from unconcentrated virions: consensus structure of prefusion S trimers
Method: subtomogram averaging / : Ke Z, Oton J, Zivanov J, Lu JM, Peukes J, Cortese M, Zila V, Scheres SHW, Briggs JAG

EMDB-11494: 
Subtomogram averaging of SARS-CoV-2 spike protein from unconcentrated virions: Prefusion Class (3 closed RBDs)
Method: subtomogram averaging / : Ke Z, Oton J, Zivanov J, Lu JM, Peukes J, Cortese M, Zila V, Scheres SHW, Briggs JAG

EMDB-11495: 
Subtomogram averaging of SARS-CoV-2 spike protein from unconcentrated virions: Prefusion Class (1 open RBD)
Method: subtomogram averaging / : Ke Z, Oton J, Zivanov J, Lu JM, Peukes J, Cortese M, Zila V, Scheres SHW, Briggs JAG

EMDB-11496: 
Subtomogram averaging of SARS-CoV-2 spike protein from unconcentrated virions: Prefusion Class (2 open RBDs)
Method: subtomogram averaging / : Ke Z, Oton J, Zivanov J, Lu JM, Peukes J, Cortese M, Zila V, Scheres SHW, Briggs JAG

EMDB-11497: 
Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: 3 Closed RBDs
Method: single particle / : Ke Z, Qu K, Nakane T, Xiong X, Cortese M, Zila V, Scheres SHW, Briggs JAG

EMDB-11498: 
Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: 2 Closed + 1 Weak RBDs
Method: single particle / : Ke Z, Qu K, Nakane T, Xiong X, Cortese M, Zila V, Scheres SHW, Briggs JAG

PDB-6zwv: 
Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: 3 Closed RBDs
Method: single particle / : Ke Z, Qu K, Nakane T, Xiong X, Cortese M, Zila V, Scheres SHW, Briggs JAG

EMDB-10835: 
High resolution cryo-EM structure of urease from the pathogen Yersinia enterocolitica
Method: single particle / : Righetto RD, Anton L, Adaixo R, Jakob R, Zivanov J, Mahi MA, Ringler P, Schwede T, Maier T, Stahlberg H

PDB-6yl3: 
High resolution cryo-EM structure of urease from the pathogen Yersinia enterocolitica
Method: single particle / : Righetto RD, Anton L, Adaixo R, Jakob R, Zivanov J, Mahi MA, Ringler P, Schwede T, Maier T, Stahlberg H

EMDB-0527: 
Chronic traumatic encephalopathy Type I Tau filament
Method: helical / : Falcon B, Zivanov J, Zhang W, Murzin AG, Garringer HJ, Vidal R, Crowther RA, Newell KL, Ghetti B, Goedert M, Scheres HW

EMDB-0528: 
Chronic traumatic encephalopathy Type II Tau filament
Method: helical / : Falcon B, Zivanov J, Zhang W, Murzin AG, Garringer HJ, Vidal R, Crowther RA, Newell KL, Ghetti B, Goedert M, Scheres HW
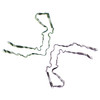
PDB-6nwp: 
Chronic traumatic encephalopathy Type I Tau filament
Method: helical / : Falcon B, Zivanov J, Zhang W, Murzin AG, Garringer HJ, Vidal R, Crowther RA, Newell KL, Ghetti B, Goedert M, Scheres HW
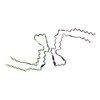
PDB-6nwq: 
Chronic traumatic encephalopathy Type II Tau filament
Method: helical / : Falcon B, Zivanov J, Zhang W, Murzin AG, Garringer HJ, Vidal R, Crowther RA, Newell KL, Ghetti B, Goedert M, Scheres HW

EMDB-0275: 
CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with picrotoxin and megabody Mb38.
Method: single particle / : Masiulis S, Desai R, Uchanski T, Serna Martin I, Laverty D, Karia D, Malinauskas T, Jasenko Z, Pardon E, Kotecha A, Steyaert J, Miller KW, Aricescu AR

EMDB-0279: 
CryoEM structure of human full-length heteromeric alpha1beta3gamma2L GABA(A)R in complex with picrotoxin, GABA and megabody Mb38.
Method: single particle / : Masiulis S, Desai R

EMDB-0280: 
CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with bicuculline and megabody Mb38.
Method: single particle / : Masiulis S, Desai R, Uchanski T, Serna Martin I, Laverty D, Karia D, Malinauskas T, Jasenko Z, Pardon E, Kotecha A, Steyaert J, Miller KW, Aricescu AR

EMDB-0282: 
CryoEM structure of human full-length heteromeric alpha1beta3gamma2L GABA(A)R in complex with alprazolam (Xanax), GABA and megabody Mb38.
Method: single particle / : Masiulis S, Desai R

EMDB-0283: 
CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with diazepam (Valium), GABA and megabody Mb38.
Method: single particle / : Masiulis S, Desai R

PDB-6hug: 
CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with picrotoxin and megabody Mb38.
Method: single particle / : Masiulis S, Desai R, Uchanski T, Serna Martin I, Laverty D, Karia D, Malinauskas T, Jasenko Z, Pardon E, Kotecha A, Steyaert J, Miller KW, Aricescu AR

PDB-6huj: 
CryoEM structure of human full-length heteromeric alpha1beta3gamma2L GABA(A)R in complex with picrotoxin, GABA and megabody Mb38.
Method: single particle / : Masiulis S, Desai R, Uchanski T, Serna Martin I, Laverty D, Karia D, Malinauskas T, Jasenko Z, Pardon E, Kotecha A, Steyaert J, Miller KW, Aricescu AR

PDB-6huk: 
CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with bicuculline and megabody Mb38.
Method: single particle / : Masiulis S, Desai R, Uchanski T, Serna Martin I, Laverty D, Karia D, Malinauskas T, Jasenko Z, Pardon E, Kotecha A, Steyaert J, Miller KW, Aricescu AR

PDB-6huo: 
CryoEM structure of human full-length heteromeric alpha1beta3gamma2L GABA(A)R in complex with alprazolam (Xanax), GABA and megabody Mb38.
Method: single particle / : Masiulis S, Desai R, Uchanski T, Serna Martin I, Laverty D, Karia D, Malinauskas T, Jasenko Z, Pardon E, Kotecha A, Steyaert J, Miller KW, Aricescu AR

PDB-6hup: 
CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with diazepam (Valium), GABA and megabody Mb38.
Method: single particle / : Masiulis S, Desai R, Uchanski T, Serna Martin I, Laverty D, Karia D, Malinauskas T, Jasenko Z, Pardon E, Kotecha A, Steyaert J, Miller KW, Aricescu AR

EMDB-4411: 
Cryo-EM structure of the human synaptic alpha1-beta3-gamma2 GABAA receptor in complex with Megabody38 in a lipid nanodisc
Method: single particle / : Laverty D, Desai R, Uchanski T, Masiulis S, Wojciech JS, Malinauskas T, Zivanov J, Pardon E, Steyaert J, Miller KW, Aricescu AR

PDB-6i53: 
Cryo-EM structure of the human synaptic alpha1-beta3-gamma2 GABAA receptor in complex with Megabody38 in a lipid nanodisc
Method: single particle / : Laverty D, Desai R, Uchanski T, Masiulis S, Wojciech JS, Malinauskas T, Zivanov J, Pardon E, Steyaert J, Miller KW, Aricescu AR

EMDB-0153: 
RELION-3.0 reconstruction of beta-galactosidase particles in EMPIAR-10061
Method: single particle / : Zivanov J, Nakane T, Scheres SHW

EMDB-0263: 
Horse spleen apo-ferritin with beam tilt correction in RELION-3.0
Method: single particle / : Zivanov J, Nakane T, Hagen WJH, Scheres SHW

EMDB-0144: 
Human apo-ferritin reconstructed in RELION-3.0
Method: single particle / : Zivanov J, Nakane T, Hagen WJH, Scheres SHW

EMDB-0152: 
RELION-3.0 reconstruction of influenza hemagglutinin (HA) trimer using particles from micrographs tilted at 40 degrees in EMPIAR-10097
Method: single particle / : Zivanov J, Nakane T, Scheres SHW
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
